
The Domino platform makes it trivial to run your analysis in the cloud on very powerful hardware (up to 32 cores and 250GB of memory), allowing massive performance increases through parallelism. In this post, we'll show you how to parallelize your code in a variety of languages to utilize multiple cores. This may sound intimidating, but Python, R, and Matlab have features that make it very simple.
Read on to see how you can get over 3000% CPU output from one machine.
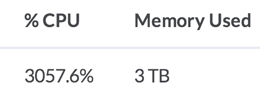
Perf stats from some parallelized Python code running on a single, 32-core machine
Is my Code Parallelizable?
For the purpose of this post, we assume a common analysis scenario: you need to perform some calculation on many items, and the calculation for one item does not depend on any other. More precisely:
- Your analysis processes a list of things, e.g., products, stores, files, people, species. Let's call this the
inputs. - You can structure your code such that you have a function which takes one such thing and returns a result you care about. Let's call this function
processInput. (After this step, you can then combine your results however you want, e.g., aggregating them, saving them to a file — it doesn't matter for our purposes.)
Normally you would loop over your items, processing each one:
for i in inputsresults[i] = processInput(i)end// now do something with resultsInstead of processing your items in a normal a loop, we'll show you how to process all your items in parallel, spreading the work across multiple cores.
To make our examples below concrete, we use a list of numbers, and a function that squares the numbers. You would use your specific data and logic, of course.
Let's get started!
Python
Python has a great package, [joblib] that makes parallelism incredibly easy.
from joblib import Parallel, delayed
import multiprocessing
# what are your inputs, and what operation do you want to
# perform on each input. For example...
inputs = range(10)
def processInput(i):
return i * i
num_cores = multiprocessing.cpu_count()
results = Parallel(n_jobs=num_cores)(delayed(processInput)(i) for i in inputsresults is now [1, 4, 9 ... ]
R
Since 2.14, R has included the Parallel library, which makes this sort of task very easy.
library(parallel)
# what are your inputs, and what operation do you want to
# perform on each input. For example...
inputs <- 1:10
processInput <- function(i) {
i * i
}
numCores <- detectCores()
results = mclapply(inputs, processInput, mc.cores = numCores)
# the above won't work on Windows, but this will:
cl <- makeCluster(numCores)
results = parLapply(cl, inputs, processInput)
stopCluster(cl)You can find some more info on the difference between mclapply and parLapply on this StackOverflow post
As an alternative, you can also use the foreach package, which lets you use a familiar for loop syntax, automatically parallelizing your code under the hood:
library(foreach)
library(doParallel)
library(parallel)
numCores <- detectCores()
cl <- makeCluster(numCores)
registerDoParallel(cl)
inputs <- 1:10
processInput <- function(i) {
i * i
}
results <- foreach(i=inputs) %dopar% {
processInput(i)
}Matlab
Matlab's Parallel Computing Toolbox makes it trivial to use parallel for loops using the parfor construct. For example:
inputs = 1:10;
results = [];
% assumes that processInput is defined in a separate function file
parfor i = inputs
results(i) = processInput(i);
endNote that if your inputs are not integers (e.g., they are file names or item identifiers), you can use the parcellfun function, which operates on cell inputs, rather than array inputs.
Octave
Unfortunately, Octave doesn't have a nice parfor equivalent — but it does have its own Parallel package. Here's how you can use it:
if exist('OCTAVE_VERSION') ~= 0
% you'll need to run this once, to install the package:
% pkg install -forge parallel
pkg load parallel
endinputs = 1:10;numCores = nproc();% assumes that processInput is defined in a separate function file[result] = pararrayfun(numCores,@processInput, inputs);Note that you can use the parcellfun function if your inputs are not numbers (e.g., if they are file names or product identifiers).
Conclusion
Modern statistical languages make it incredibly easy to parallelize your code across cores, and Domino makes it trivial to access very powerful machines, with many cores. By using these techniques, we've seen users speed up their code over 32x while still using a single machine.
If you'd like to try applying this approach to your analysis, please let us know, we're happy to help!
Nick Elprin is the CEO and co-founder of Domino Data Lab, provider of the open data science platform that powers model-driven enterprises such as Allstate, Bristol Myers Squibb, Dell and Lockheed Martin. Before starting Domino, Nick built tools for quantitative researchers at Bridgewater, one of the world's largest hedge funds. He has over a decade of experience working with data scientists at advanced enterprises. He holds a BA and MS in computer science from Harvard.
Subscribe to the Domino Newsletter
Receive data science tips and tutorials from leading Data Science leaders, right to your inbox.
By submitting this form you agree to receive communications from Domino related to products and services in accordance with Domino's privacy policy and may opt-out at anytime.



