Speeding up Machine Learning with parallel C/C++ code execution via Spark
Nikolay Manchev2022-02-16 | 12 min read
The C programming language was introduced over 50 years ago and it has consistently occupied the most used programming languages list ever since. With the introduction of the C++ extension in 1985 and the addition of classes and objects, the C/C++ pair keep a central role in the development of all major operating systems, databases, and performance critical applications in general. Because of its efficiency, C/C++ underpin a large number of machine learning libraries (e.g. TensorFlow, Caffe, CNTK) and widely used tools (e.g. MATLAB, SAS). C++ may not be the first thing that springs to mind when thinking about Machine Learning and Big Data, but it is omnipresent everywhere in the field where lightning fast computations are needed - from Google's Bigtable and GFS to pretty much everything GPU related (CUDA, OCCA, openCL etc.)
Unfortunately, when it comes to parallelised data processing, C/C++ do not tend to provide out-of-the-box solutions. This is where Apache Spark, the king of large-scale data processing, comes to the rescue.
Calling C/C++ via RDD.pipe
According to its documentation, the pipe operator enables Spark to process RDD data using external applications. The use of pipe is not C/C++ specific, and it can be used to call external code written in an arbitrary language (shell scripts included). Internally, Spark uses a special type of RDD, the PipedRDD, to pipe the contents of each data partition through the specified external program.
Let's look at a simple example. Here is a basic C program that reads from the standard input (stdin) and parrots back the string, prefixed with a "Hello". The program is stored in a file named hello.c
/* hello.c */
#include <stdio.h>
#include <stdlib.h>
int main(int argc, char *argv[]) {
char *buffer = NULL;
int read;
size_t len;
while (1) {
read = getline(&buffer, &len, stdin);
if (-1 != read)
printf("Hello, %s", buffer);
else
break;
}
free(buffer);
return 0;}
Let's compile and test it. Note that you can press Ctrl+D to interrupt the execution.
$ gcc -o hello hello.c
$ ./hello
Dave
Hello, Dave
KikoHello, Kiko
DirkHello, Dirk
$So far so good. Now let's see how we can pipe this code to a Spark RDD. First, there are two things that we'll need to make sure are in place before we try to actually execute the compiled C code:
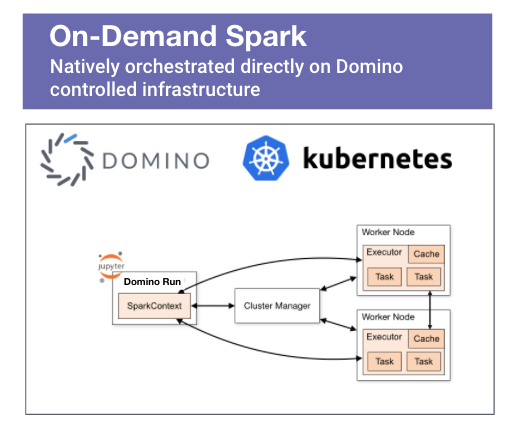
- Access to a Spark cluster - this is an obvious pre-req and spinning up an on-demand Spark cluster in the Domino MLOps platform is a straightforward operation that is completed with just a few clicks. Alternatively, you could run Spark in standalone mode.
- The external code that we are planning on using should be available to all Spark executors tasked with running the application. If we are using Domino on-demand Spark, all executors have default access to a shared dataset. All we have to do is place the compiled C/C++ program in the dataset's folder. Alternatively, we can modify the driver program to include a call to SparkContext.addFile(), which will force Spark to download the external code referenced by the job on every node. We'll show how to handle both of the scenarios outlined above.
Assuming that we are using Domino, we can now place the compiled hello program into a dataset. If the name of our project is SparkCPP, then by default we'll have access from within our Workspace to a dataset located at /domino/datasets/local/SparkCPP/. Let's copy the hello program there.
$ cp hello /domino/datasets/local/SparkCPP/
Next, we create the main program test_hello.py, which we'll use to test the piping. This PySpark application will simply parallelise an RDD and directly pipe the external code located inside the dataset.
from pyspark.sql import SparkSession
spark = SparkSession \
.builder \
.appName("MyAppName") \
.getOrCreate()
# Suppress logging beyond this point so it is easier to inspect the output
spark.sparkContext.setLogLevel("ERROR")
rdd = spark.sparkContext.parallelize(["James","Cliff","Kirk","Robert", "Jason", "Dave"])
rdd = rdd.pipe("/domino/datasets/local/SparkCPP/hello")
for item in rdd.collect():
print(item)
spark.stop()
For a remote cluster (i.e. a cluster that is independent and is not running in Domino) we can modify the code to explicitly distribute the hello program. Notice that the access path for the pipe command changes, as the hello code is now in the working directory of each executor. We are also assuming that the absolute path of the compiled C program is /mnt/hello.
from pyspark.sql import SparkSession
spark = SparkSession \
.builder \
.appName("HelloApp") \
.getOrCreate()
spark.sparkContext.addFile("/mnt/hello")
rdd = spark.sparkContext.parallelize(["James","Cliff","Kirk","Robert", "Jason", "Dave"])
rdd = rdd.pipe("./hello")
for item in rdd.collect():
print(item)
spark.stop()
Running the program via spark-submit yields the same output in both cases:
$ spark-submit test_hello.py
2022-02-10 09:55:02,365 INFO spark.SparkContext: Running Spark version 3.0.0
…
Hello, James
Hello, Cliff
Hello, Kirk
Hello, Robert
Hello, Jason
Hello, Dave
$ We see that our C program correctly concatenates all entries of the RDD. Moreover, this process is executed in parallel on all available workers.
Using compiled libraries and UDFs
Now let's move to something more advanced. We'll see how to use a compiled library (shared library or a collection of shared objects, if we want to use the Linux term) and map a function from it to a Spark User Defined Function (UDF).
We start by placing a simple C function in a file named fact.c that computes the factorial of a number n:
/* fact.c */
int fact(int n) {
int f=1;
if (n>1) {
for(int i=1;i<=num;i++)
f=f*i;
}
return f;
}The code above is pretty self-explanatory. However, instead of calling it directly like the hello example, we will compile it to an .so library and define a UDF wrapper around fact(). Before we do that though, we need to consider what options are out there that can help us with interfacing Python and C/C++.
- SWIG (Simplified Wrapper and Interface Generator) - a mature framework that connects programs written in C and C++ with a variety of high-level programming languages like Perl, Python, Javascript, Tcl, Octave, R and many others. Originally developed in 1995, SWIG is a mature and stable package, the only downside being that its flexibility comes at a price - it is complex and requires a number of additional pre-compilation steps. It is worth noting, that because SWIG is more or less language agnostic, you can use it to interface Python to any language supported by the framework. Its capability to automatically wrap entire libraries (provided we have access to the headers) is also quite handy.
- CFFI (C Foreign Function Interface) - according to its documentation, the main way to use CFFI is as an interface to some already-compiled shared object which is provided by other means. The framework enhances Python with a dynamic runtime interface, which makes it easy to use and integrate. The runtime overhead, however, leads to a performance penalty so it lags behind statically compiled code.
- Cython - a superset of Python, Cython is a compiled language designed for speed, which provides additional C-inspired syntax to Python. Compared to SWIG, the learning curve of Cython is less steep, because it automatically translates Python code to C, so programmers don't have to implement logic in lower-level languages to get a performance boost. On the downside, Cython requires an additional library at build time and has a separate installation, which complicates the deployment process.
Of course, there are other options out there (Boost.Python and Pybindgen spring to mind), but a comprehensive overview of every C/C++ interfacing tool is out of the scope of this text. For now, we'll provide an example of using SWIG, which is more or less our favourite because of its maturity and versatility.
The installation of SWIG on Linux cannot be any simpler - it boils down to a single sudo apt install swig command. If you want to see the installation process for other operating systems, please feel free to look at its documentation. There is also a simple tutorial that provides a 10 minute "getting started" introduction and is an easy way for everyone who wants to get familiar with using the framework.
Assuming that we have SWIG installed, the first step required to build the shared library is to create an interface for it. This file serves as the input to SWIG. We will name the file for our fact() function fact.i (i stands for interface) and we'll put the function definition inside like so:
/* fact.i */
%module fact
%{
extern int fact(int n);
%}
extern int fact(int n);
We then run SWIG to build a Python module.
$ swig -python fact.iWe see that SWIG generates two new files:
- fact.py (based on the module name in the interface file) is a Python module that we can import directly
- fact_wrap.c file, which should be compiled and linked with the rest of the external code
$ ls -lah
total 132K
drwxrwxr-x 2 ubuntu ubuntu 4.0K Feb 10 14:32 .
drwxr-xr-x 6 ubuntu ubuntu 4.0K Feb 10 11:13 ..
-rw-rw-r-- 1 ubuntu ubuntu 110 Feb 10 11:20 fact.c
-rw-rw-r-- 1 ubuntu ubuntu 55 Feb 10 14:32 fact.i
-rw-rw-r-- 1 ubuntu ubuntu 3.0K Feb 10 14:32 fact.py
-rw-rw-r-- 1 ubuntu ubuntu 112K Feb 10 14:32 fact_wrap.c
$ Next, we use GCC to compile the function and the wrapper:
$ gcc -fpic -I /usr/include/python3.6m -c fact.c fact_wrap.c
$Finally, we bundle the object files into a shared library named _fact.so:
$ gcc -shared fact.o fact_wrap.o -o _fact.so
$ ls -lah
total 240K
drwxrwxr-x 2 ubuntu ubuntu 4.0K Feb 10 14:36 .
drwxr-xr-x 6 ubuntu ubuntu 4.0K Feb 10 11:13 ..
-rw-rw-r-- 1 ubuntu ubuntu 108 Feb 10 14:33 fact.c
-rw-rw-r-- 1 ubuntu ubuntu 89 Feb 10 14:35 fact.i
-rw-rw-r-- 1 ubuntu ubuntu 1.3K Feb 10 14:35 fact.o
-rw-rw-r-- 1 ubuntu ubuntu 3.0K Feb 10 14:35 fact.py
-rwxrwxr-x 1 ubuntu ubuntu 51K Feb 10 14:36 _fact.so
-rw-rw-r-- 1 ubuntu ubuntu 112K Feb 10 14:35 fact_wrap.c
-rw-rw-r-- 1 ubuntu ubuntu 50K Feb 10 14:35 fact_wrap.o
$We now need to put the shared library in a place that will be accessible from all worker nodes. The options are more or less the same - a shared file system (e.g. Domino dataset) or add the library during execution via SparkContext.addFile().
This time, we'll use a shared dataset. Don't forget to copy both the compiled library and the Python wrapper classes to the shared file system.
$ mkdir /domino/datasets/local/SparkCPP/fact_library
$ cp _fact.so /domino/datasets/local/SparkCPP/fact_library/
$ cp fact.py /domino/datasets/local/SparkCPP/fact_library/Here is the Spark application that we'll use to test the external library call.
import sys
from pyspark.sql import SparkSession
from pyspark.sql.functions import udf
from pyspark.sql.types import IntegerType
def calculate_fact(val):
sys.path.append("/domino/datasets/local/SparkCPP/fact_library")
import fact
return fact.fact(val)
factorial = udf(lambda x: calculate_fact(x))
spark = SparkSession \
.builder \
.appName("FactApp") \
.getOrCreate()
df = spark.createDataFrame([1, 2, 3, 4, 5], IntegerType()).toDF("Value")
df.withColumn("Factorial", \
factorial(df.Value)) \
factorial(df.Value)) \
.show(truncate=False)
spark.stop()There are a couple of things to notice here. First, we are using calculate_fact() as a User Defined Function (UDF). UDFs are user-programmable routines that act on a single RDD row and are one of the most useful features of SparkSQL. They allow us to extend the standard functionality of SparkSQL and promote code reuse. In the code above we use udf() from the org.apache.spark.sql.functions package, which takes a Python function and returns a UserDefinedFunction object. The Python function above, however, is a call to our shared library and the underlying C implementation. The second thing to notice is that we use sys.path.append(), which tells Python to add a specific path for the interpreter to search when loading modules. We do this to force python to look for the fact module located under /domino/datasets/local/SparkCPP/fact_library. Notice that the append() and import calls are part of the calculate_fact() function. We do it this way because we want the path appended and the module imported on all worker nodes. If we bundle these with the other imports at the top of the script, the import will only happen at the Spark Driver level and the job will fail.
Here is the output from running the Spark application above. :
$ spark-submit test_fact.py
2022-02-11 16:25:51,615 INFO spark.SparkContext: Running Spark version 3.0.0
…
2022-02-11 16:26:07,605 INFO codegen.CodeGenerator: Code generated in 17.433013 ms
+-----+---------+
|Value|Factorial|
+-----+---------+
|1 |1 |
|2 |2 |
|3 |6 |
|4 |24 |
|5 |120 |
+-----+---------+
…
2022-02-11 16:26:08,663 INFO util.ShutdownHookManager: Deleting directory /tmp/spark-cf80e57a-d1d0-492f-8f6d-31d806856915
$
Summary
In this article, we looked at how Spark, the undisputed king of big data processing, can be enhanced with access to lightning-fast operations provided by external C/C++ libraries. Note, that this approach is not exclusively limited to C and C++, as frameworks like SWIG provide access to many other languages like Lua, Go, and Scilab. Other projects like gfort2py also provide access to Fortran.
Moreover, one could use NVIDIA CUDA to write, compile and run C/C++ programs that both call CPU functions and launch GPU kernels. Such code can then be accessed through Spark via shared libraries, providing both parallelization and GPU-accelerated custom operations.
Additional resources
- The Domino Enterprise MLOps platform provides flexible access to on-demand Spark compute. It offers the ability to dynamically provision and orchestrate a Spark cluster directly on the cloud or on-prem infrastructure backing the Domino instance. It can also run containers directly from NVIDIA's NGC catalog. Sign up for and see how the platform can help you accelerate model velocity in your organization.
- Learn more about Spark, Ray, and Dask and how to select the right framework for your machine learning pipeline.
Nikolay Manchev is the Principal Data Scientist for EMEA at Domino Data Lab. In this role, Nikolay helps clients from a wide range of industries tackle challenging machine learning use-cases and successfully integrate predictive analytics in their domain specific workflows. He holds an MSc in Software Technologies, an MSc in Data Science, and is currently undertaking postgraduate research at King's College London. His area of expertise is Machine Learning and Data Science, and his research interests are in neural networks and computational neurobiology.
RELATED TAGS
Subscribe to the Domino Newsletter
Receive data science tips and tutorials from leading Data Science leaders, right to your inbox.
By submitting this form you agree to receive communications from Domino related to products and services in accordance with Domino's privacy policy and may opt-out at anytime.



